An R-loop reorganizes a small section of a chromosome: DNA strands are separated and one of them is bound to its complementary RNA sequence. They appear frequently during elongation, and for many genes, they are essential for their proper expression. However, exposed single DNA strand breaks easily, and sometimes the second strand breaks too. When DNA of Ig genes is permuted to form specific antibodies, R-loops assist in making double DNA breaks that are necessary for this process. In other cases, DNA repair pathways heal the breaks. But repairs may fail, causing various diseases, especially cancers and neurodegenerative diseases.
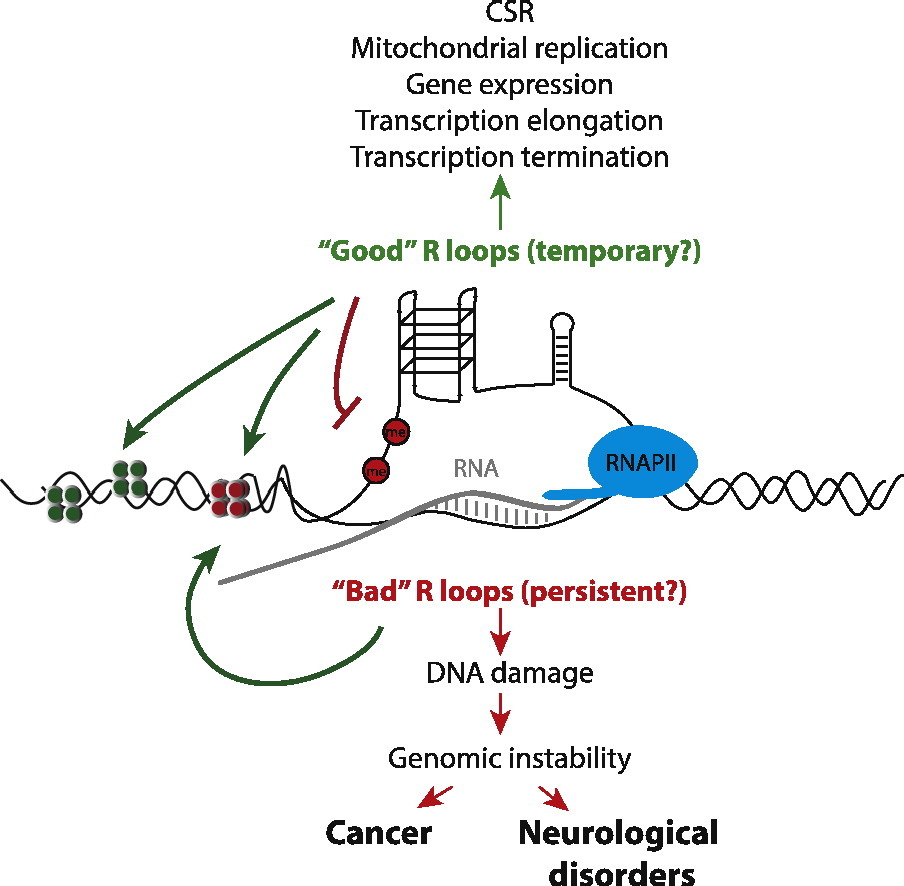
One may ask: where the complementary RNA comes from? They are untranscribed fragments of pre-mRNA, introns and parts of UTRs. R-loops are typically dismantled, RNases degrade the RNA fragment and the two DNA fragments hybridize which restores the double helix. However, a persistent R-loop in 5’UTR may be required for proper transcription initiation, and an R-loop in 3’UTR stops elongation more efficiently. There exists diseases caused by missing of such persistent R-loops.
Importance of R-loops was discovered only in 2011, but now they are studied very widely, e.g. 20 publications found by PubMed for July 2023. Accumulated knowledge on R loops gives more complete explanations to many important questions. Example 1: cancers are caused in part by chromosomal breaks, why cancers of different origins have different hotspots of those breaks? Persistent R-loops are related to gene expression, so they differ for different cell types. Example 2: why deficiency in FA DNA repair pathway changes phenotypes even when “better” DNA repair pathways function? FA pathway is essential for repair of breaks caused by R-loops. And there are many other genes that are essential for R-loop regulation.
Patricia Richard and James L. Manley, R Loops and Links to Human Disease, J Mol Biol (2017) 429, 3168–3180

