Interpreting gene expression and epigenetic data requires understanding heterochromatin assembly, regulatory mechanisms that can silence gene expression in wider chromosomal regions, and when those mechanisms are blocked, we can observe activation of gene expression. Shiv Greval reviews new discoveries and gives pointers to 204 references in “The molecular basis of heterochromatin assembly and epigenetic inheritance”, Molecular Cell, May 18, 2023. The focus is on facultative heterochromatin, marked by H3K9me that silences genes, mostly in a lineage specific manner.
By necessity, some new findings are described very tersely, so one may need to check references to understand them, but key new findings are described in detail and illustrated.
For example, heterochromatin assembly can be initiated by sequence specific binding of a chromatin complex that includes a transcription factor, and then spreads across a domain that in both directions is flanked by a boundary element and an active transcription starting site (marked with H3K4me3). This is not exactly new, but now we know three distinct mechanisms that form such silenced domains, usually tethered to nuclear lamina.
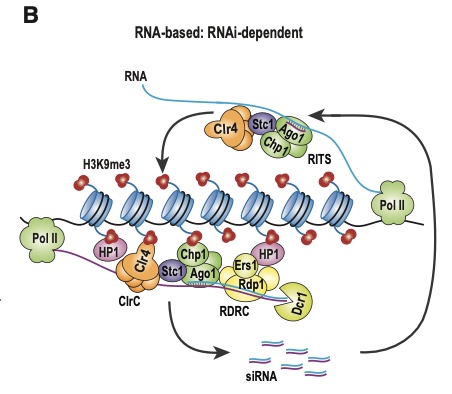
One mechanism connects to expression of microRNA genes that are embedded in protein coding genes: a protein coding transcript is processed by an RNA interference complex that leaves only siRNA sequences included in this transcript and degrades the rest. In turn, those siRNA sequences participate in the recruitment of chromatin factors needed in heterochromatin assembly (this is illustrated in the figure of this post). This may help interpreting differential expression of genes with “RNA processing” GO term and expression of miRNA genes. I must admit that so far, I did not know how to interpret such genes.